The challenge of using AI for non-traditional data modelling in predictive analytics
Authors: Justin Green, Luke Farrow, Feroz Dinah and Vipin Asopa
The challenge of using AI for non-traditional data modelling in predictive analytics
Justin Green has recently completed his Clinical Research Fellowship with NAHR, BHS and ORUK, exploring the application of digital technologies in determining longitudinal outcomes in hip preservation surgery. He is currently reading for a PhD in Artificial Intelligence at Newcastle University with an interest in ethics and responsible application of AI in healthcare. Justin acts a Clinical Safety Officer and member of the AI Clinical Safety Board at Northumbria Healthcare NHS Foundation Trust and contributes to the NortHFutures Digital Skills collaborative.
Luke Farrow is a Scottish Research Excellence Development Scheme (SCREDS) Clinical Lecturer based in Aberdeen. He is currently in the final stages of a Clinical Academic Fellowship exploring how to make improvements in the clinical care pathway for those awaiting hip and knee replacement through the use of AI. He leads the Aberdeen AI Healthcare Collaborative (AAHC) and is the current BOA Associate Sub-Speciality Lead for Elective Orthopaedics. Luke has particular research interests in the use of routinely collected healthcare information and clinical applications of AI in T&O.
Feroz Dinah is a Consultant Hip and Knee Surgeon based in the Epsom and St Helier Hospitals NHS Trust. He also works closely with the research department in the South West London Elective Orthopaedic Centre. He has a keen interest in the application of robotic surgery and AI in trauma and orthopaedics.
Vipin Asopa is a Specialist Hip and Knee Surgeon at South West London Elective Orthopaedic Centre, Epsom. His research interests include the use of artificial intelligence to improve patient outcomes following surgery.
Much has been written about the use of predictive analytics in orthopaedics in terms of predicting outcomes of surgery, optimising operational efficiency, etc1. Predictive analytics has classically been applied to ‘traditional’ datasets such as tabular continuous or discrete data in order to identify trends and relationships. However, artificial intelligence (AI) can also handle ‘non-traditional’ data such as unfiltered social media text and images through natural language processing (NLP) and computer vision (CV), to reach a predictive output. However, explaining how AI reaches such predictions can be problematic, especially with complex setups like convoluted neural networks (CNNs) and graphical neural networks (GNNs). Such ‘black box’ analytics are opaque and pose challenges in relation to data quality and output, privacy, trusting the output and ethical considerations2.
This article will start with a description of using predictive analytics with non-traditional (NT) data. It will then discuss the model complexity vs. explainability to optimise accuracy and user confidence. Lastly, the importance of reporting guidelines regarding predictive analytics in healthcare will be discussed.
Predictive analytics and non-traditional data
NT data refer to data that cannot be easily handled by traditional statistical tools or methods because the data does not fit neatly into the fields, or it may be too unstructured or varied to fit into a traditional database. The data may also be too large: examples include the billions of search engine uses undertaken on a daily basis, resulting in huge amounts of information. Such data volumes may be measured in Petabytes to Exabytes (1,000 to 1 million TB) and is typically termed ‘Big data’.
Machine learning (ML) allows big data to be processed in a stream, i.e. analysed and acted upon in near real-time, rather than being collected and stored for later batch processing because of the above challenges. Other examples of challenging data include newer forms of personal data produced by various connected digital platforms such as social media posts, or devices like smartphones, fitness trackers, computer tablets, smart home devices, medical devices, etc., resulting in large amounts of unstructured text, numerical and image data. Because this type of data is not systematically structured or stored, its analysis has only become possible thanks to advances in AI and redictive analytics. Previous analysis of large amounts of data, such as that found in the Facebook and Instagram ecosystems relied on traditional statistical methods and simple algorithms, requiring human intervention. These were far less efficient than modern AI-based techniques such as CNNs and more recently, GNNs, as discussed below.
Graph data is another form of NT data. It is a non-linear data structure representing relationships between entities (Figure 1).
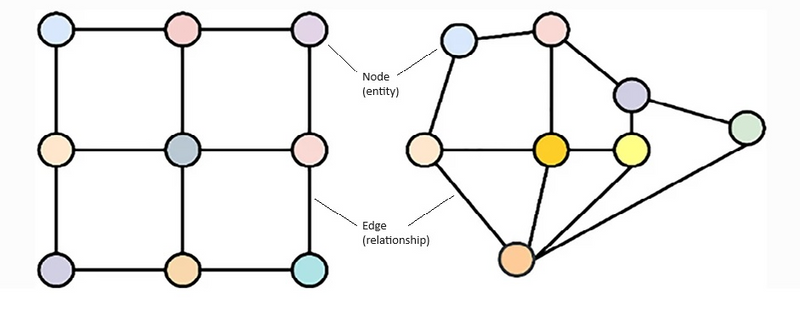
Figure 1: Diagrammatic representation of graph data. Graphs lack a predefined structure for data storage (left side), and there is no inherent knowledge of node-to-neighbour relationships, as illustrated on the right side. Left side is amenable to CNNs, right side requires GNNs.
The most common applications include social networks (connections between friends or followers) and recommendation systems (personalised suggestions while the customer is still browsing). Graph data can also be used to describe group dynamics in a team sport, to find the shortest route in a navigation programme, and to identify spread of information or even diseases among social networks.
The presence of different interconnected data sources, found in various domains, from social networks, recommendation systems, science and cybersecurity, has fuelled the rapid evolution of GNNs3. These networks can model and understand complex relationships and make sense of the interlinked data to help solve real-world problems better than traditional ML models (e.g. CNNs).
Deep learning (DL), a subset of ML, is a powerful method that can be used to analyse the unstructured NT data described above (see Figure 2).
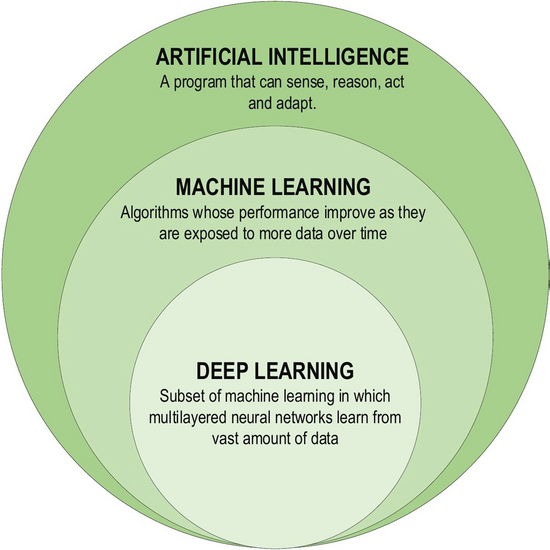
Figure 2: Deep learning as a subset of machine learning (from AlZubaidi et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data. 2021;8,:53).
It is based on artificial neural networks using multiple layers that simulate the human brain3. NLP and CV are applications of DL that help with data analysis. Examples of NLP include language translation and speech recognition, while medical applications include the analysis of clinical, patient communication and research summarisation, if necessary, after conversion from image or sound data to textual data1. Similarly, CV examples include object detection (autonomous surveillance), image classification (medical radiology, dermatology and pathology) and face recognition, with orthopaedic applications including implant positioning and gait analysis1.
Although DL algorithms perform best after being developed/trained on large datasets, they require significant computational resources including specialised hardware4. Furthermore, DL needs vast amounts of labelled training data, and the performance of a trained algorithm depends heavily on how the training data represents the data being analysed. If the dataset is small or noisy, the model can overfit to the training data and will not be applicable to unseen data. This can cause models to learn and propagate biases present in the training data, leading to unfair or discriminatory outcomes. This can raise ethical questions, particularly around privacy, consent, and fairness. DL models are often criticised for their lack of transparency and interpretability and explainability, making it difficult to understand how decisions are made5. This is addressed below.
Making AI models less opaque: Explainable AI (XAI)
The term ‘black box’, originally coined during World War II to describe aircraft components housing sensitive data, was first applied to AI in 1961, to denote analysis or interpretation through unknown means. More recently, the term has evolved to signify the opaque nature of ML models6. ML and DL models, construct non-linear relationships in data. This enables more complexity and in turn results higher accuracy than traditional statistical models. The complexity of the relationships however generates a lack of clarity in the modelling structure. When the arrays are interrogated as to which factors in training data leads to the particular conclusions it can become too complex to interpret. This opacity has earned them the name ‘Black Boxes’, highlighting the difficulty in understanding and interpreting their inner workings. Lack of transparency raises trust and accountability concerns with users, especially in critical applications such as healthcare.
To address this, Explainable Artificial Intelligence (XAI) proposes a shift from the black box model to a fully transparent AI by devising methods and tools to mitigate the opacity in models, without degrading their accuracy and performance. This, it is hoped, would make the judgments of complex models understandable and expected, by revealing the internal mechanisms in a comprehensible manner7.
XAI can be represented in various ways, from mathematical equations to visual representations such as scatter plots. An example of XAI in medical imaging is employing saliency mapping, a process where areas of an image a model considers important in generating a prediction, are highlighted providing a visual indication of the regions of interest which are determining the outcomes. The use of saliency mapping as a tool, has shown promising results in implant identification and models predicting the risk of implant failure or loosening8 improving the interpretation features. Similarly, LIME (Local Interpretable Model-agnostic Explanations) approximates complex models with simpler, interpretable models locally around a prediction, offering insights into individual decisions. SHAP (SHapley Additive exPlanations) values, based on cooperative game theory, provide a unified measure of feature importance in predictive analytics, making it possible to understand the contribution of each feature in the data to the model’s output.
Generative AI, seen in LLMs, has been used to create new content such as text, images, and sound, also faces significant challenges regarding explainability. In the context of orthopaedics, generative AI has been used to simulate and visualise surgical outcomes in arthroplasty. For example, a group led by Bardia Khosravi from the Mayo Clinic9 applied Generative Adversarial Networks (GANS) to create high-fidelity synthetic pelvis radiographs which were used for DL-based image analysis. The synthetic images were indistinguishable from real images and showed equivalent performance when assessed by DL models. However, the use of synthetically generated images to train other algorithms raises questions of whether this is a suitable way to train algorithms and, in turn, the validity of the decision making that arises from these. This makes the results difficult to interpret. The challenge lies in understanding how these models generate specific outputs from the given inputs, particularly when the training data may contain inherent broad variation in structure and biases. A systemic review reported that the diagnostic accuracy of LLMs was significantly worse than clinicians10. To ensure that patient safety in an autonomous clinical decision-making scenario is maintained, transparency and assurances in LLMs is vital, and assurances in performance should be validated to a level that is at least as good as, if not better, than clinicians.
The lack of overall transparency with how AI algorithms work is a barrier to humans trusting and accepting these tools. It highlights the need for a robust approach to the development, training, testing and reporting of AI algorithms in healthcare.
Reporting guidelines for predictive analytics in orthopaedics
One of the other key advancements helping drive improvements around the potential clinical integration of AI based predictive analytics is the formation of associated development and reporting guidelines.
The Clinical Practice Integration of AI (CPI-AI) framework11 is one proposed application using the IDEAL principles12 of surgical innovation to AI applications in T&O. It identifies the steps required from the beginning of an AI based research proposal through to eventual clinical deployment and includes six stages:
• Stage 0 – Concept outline
• Stage 1 – Algorithm development
• Stage 2a – External validation
• Stage 2b – Prospective assessment
• Stage 3 – Clinical impact assessment (Randomised Controlled Trial)
• Stage 4 – Implementation and model surveillance
The majority of current work in T&O using AI based predictive analytics falls into stage 1, with very few progressing beyond this to build the evidence base for necessary regulatory approvals and eventual clinical practice integration13. The application of NLP for prediction of selection for hip and knee arthroplasty surgery has previously demonstrated the importance of external validation in the accurate assessment of predictive capability, given a significant drop in model performance when tested on new external data sources14.
Another key aspect to the development of AI based predictive analytics is the use of reporting checklists. These serve not only as guidance to researchers who are developing AI algorithms, but also provide for a robust system of assessment of quality and diligence. Several reporting checklists have been developed specifically for AI applications for various research methodology, including predictive analytics15-18.
The main checklist in this regard is the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis AI (TRIPOD+AI) statement15, which was published by Collins et al. following a modified Delphi process. This checklist establishes 27 criteria across the domains of study title, abstract, introduction, methods, results and discussion.
One of the key changes to the original TRIPOD guidance is an emphasis on the importance of fairness in the evaluation of AI. Points considered include evaluation of model performance based on key subgroups (for example different sociodemographic profiles which will vary dependent on the target application), as well as reporting standards that include patient and public involvement, and open science principles. The development of the associated Prediction model Risk Of Bias ASsessment Tool (PROBAST+AI) is still underway and will further help delineate assessment of the quality and risk of bias in prediction models16.
Other checklists such as the Developmental and Exploratory Clinical Investigations of DEcision support systems driven by Artificial Intelligence (DECIDE-AI)17 and Consolidated Standards of Reporting Trials-Artificial Intelligence (CONSORT-AI)18 statements are also important in relation to further improving and reporting predictive analytics, particularly as a research proposal moves towards clinical practice integration (CPI-AI stages 2b and 3). The DECIDE-AI checklist particularly focuses on early-stage clinical evaluation of AI systems, including predictive analytics and includes 17 specific reporting items with a particular focus on proof of clinical utility, safety, and human factors, in preparation for large-scale trials. The final consideration is the CONSORT-AI statement that covers the conduct of Randomised Controlled Trials related to AI interventions and adds a further 14 new checklist items to the original CONSORT statement.
These reporting standards, when added to the CPI-AI framework, can provide a clear pathway for safe and evidence-based development and deployment of predictive AI in T&O.
Conclusion
This article has highlighted the challenges around analysing the huge amount of NT data produced by personal and other electronic devices which could help with patient care. Newer methods of AI, capable of dealing with such huge amounts of varied data have been described. Safeguards about using and reporting the use of AI models in healthcare have been explained. The importance of these cannot be underestimated to help improve the quality of research and assist the future integration of predictive AI into clinical practice.
References
- Coppola A, Hing C, Asopa V. Integrating artificial intelligence into trauma and orthopaedics: History, current state of AI in T&O and future perspectives. Orthopaedics Online, May 2024. Available at: www.boa.ac.uk/resource/integrating-artificial-intelligence-into-trauma-and-orthopaedics.html.
- Coppola A, Asopa V. A practical approach to AI in T&O. Journal of Trauma and Orthopaedics. 2024;12(2):30-2.
- Kriegeskorte N, Golan T. Neural network models and deep learning. Current Biology. 2019;29:R231-R236
- AlZubaidi L, Zhang J, Amjad HJ, et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data. 2021;8:53.
- Samek W, Wiegand T, Müller K-R. Explainable Artificial Intelligence: Understanding, Visualizing and Interpreting Deep Learning Models. 2017. arXiv preprint arXiv:1708.08296.
- Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nature Machine Intelligence. 2019;1(5):206-15.
- Hassija V, Chamola V, Mahapatra A, et al. Interpreting Black-Box Models: A Review on Explainable Artificial Intelligence. Cogn Comput. 2024;16:45-74.
- Khemani B, Patil S, Kotecha K. et al. A review of graph neural networks: concepts, architectures, techniques, challenges, datasets, applications, and future directions. J Big Data. 2024;11:18.
- Khosravi B, Rouzrokh P, Mickley JP, et al. Creating High Fidelity Synthetic Pelvis Radiographs Using Generative Adversarial Networks: Unlocking the Potential of Deep Learning Models Without Patient Privacy Concerns. J Arthroplasty. 2023;38(10):2037-43.e1.
- Hager P, Jungmann F, Holland R, et al. Evaluation and mitigation of the limitations of large language models in clinical decision-making. Nat Med. 2024 Jul 4. Epub ahead of print.
- Farrow L, Meek D, Leontidis G, et al. The Clinical Practice Integration of Artificial Intelligence (CPIAI) Framework - an Adaptation of the IDEAL Approach to Artificial Intelligence Applications in Trauma and Orthopaedics. Bone and Joint Research. In Press.
- McCulloch P, Altman DG, Campbell WB, et al. No surgical innovation without evaluation: the IDEAL recommendations. Lancet. 2009;374(9695):1105-12.
- Dijkstra H, van de Kuit A, de Groot T, et al. Systematic review of machine-learning models in orthopaedic trauma. Bone Jt Open. 2024;5(1):9-19.
- Farrow L, Zhong M, Anderson L. Use of natural language processing techniques to predict patient selection for total hip and knee arthroplasty from radiology reports. Bone Joint J. 2024;106-B(7):688-95.
- Collins GS, Moons KGM, Dhiman P, et al. TRIPOD+AI statement: updated guidance for reporting clinical prediction models that use regression or machine learning methods. BMJ 2024;385:e078378.
- Collins GS, Dhiman P, Andaur Navarro CL, et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open. 2021;11(7):e048008.
- DECIDE-AI Steering Group. DECIDE-AI: new reporting guidelines to bridge the development-to-implementation gap in clinical artificial intelligence. Nat Med;27(2):186-7.
- Liu X, Cruz Rivera S, Moher D, et al. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat Med. 2020;26:1364-74.
